Optimization with PyGMO
The following paragraphs will give some information on how to optimize an astrodynamics problem written with tudatpy through the usage of PyGMO. The aim of this page is not to provide comprehensive documentation to the usage of PyGMO (such a guide already exists, see previous link), but rather to introduce the reader to the logic behind PyGMO and illustrate how to employ it jointly with tudatpy.
Content of this page
About PyGMO
The “basic” idea
PyGMO is a Python scientific library derived from PaGMO (Parallel Global Multiobjective Optimization), an open-source software developed at the European Space Agency by F. Biscani and D. Izzo [Biscani2020]. The flexible and complete framework of PaGMO (and of its equivalent PyGMO) can be applied to “single-objective, multiple-objective, continuous, integer, box-constrained, non linear constrained, stochastic, deterministic optimization problems”. Both programs are based on the concept of the island model, which in short is a computational strategy that allows to run the optimization process in parallel (hence its name) by using multithreading techniques.
Installing PyGMO
PyGMO is not part of the standard tudatpy distribution, but it can easily be added to any tudatpy environment.
Here we will assume that you already have a standard tudatpy environment (called tudat-space) installed.
If that is not the case, please follow the “Installing Tudat(Py)” instructions in Installation.
In order to make PyGMO available inside the tudat-space environment, the following steps need to be taken.
First, activate the tudat-space environment:
$ conda activate tudat-space
Next, use conda to install the PyGMO package to this environment:
(tudat-space) $ conda install pygmo
Warning
Please ensure to install PyGMO in your given tudatpy environment. Do not add the PyGMO package to your base environment.
Note
If adding pygmo to an existing conda environment returns errors, create a new conda environment with pygmo added as a dependency.
name: tudat-space
channels:
- conda-forge
- tudat-team
dependencies:
- tudatpy
- matplotlib
- scipy
- pandas
- jupyterlab
- pygmo # <-- Add this line to your environment.yaml
Lastly, you should verify that the package is now available in your tudatpy environment. You can do so via your IDE or by using the following command in your terminal:
(tudat-space) $ conda list | grep pygmo
which should return the name, version, build and channel of the pygmo installation in the given environment.
First steps
There are a number of basic elements concurring to the solution of an optimization problem using PyGMO. These will be listed below and briefly explained; each of them correspond to an equivalent base class in PyGMO.
1. A problem. This represents the problem for which an optimal solution must be found. This is usually known, in PyGMO terminology, as User-Defined Problem (UDP); it is usually more interesting to code and solve a custom problem, but there are certain problems that are defined within PyGMO and can be readily used.
2. An algorithm. This is the procedure to solve the optimization problem. Similarly, this is known as User-Defined Algorithm (UDA); differently from the problem, it is often more convenient to take advantage of the many UDAs offered by PyGMO. Nonetheless, it is always possible to code a custom solver.
3. One (or more) individuals. Optimizers rely not on one, but many decision vectors that may interact (heuristic optimizers) or may not (analytical solvers) interact with each other. A set of individuals form a population. In PyGMO, it is more frequent to deal with an entire population, rather than with a single individual.
There are also two other fundamental blocks that conclude the structure of the island model. One is the island itself, which represents the main parallelization block to achieve simultaneous computation; the other one is the archipelago, consisting of a set of islands. These are useful to solve more advanced and computationally-demanding problems, therefore their usage will not be analyzed in detail here.
Optimizing a simple problem
In this example, we will attempt to optimize a simple problem: the minimization of a known analytical function. We chose Himmelblau’s function as it is often employed to test the performance of optimization algorithms:
subject to the bounds:
There are four equal minima which can be found analytically. These are:
Below, we will explain how to write the code for this specific UDP and solve it with PyGMO.
The original code, which is broken down into parts for the sake of clarity, is available here.
1. Creation of the UDP class
First, we create a Python class to describe the problem. This class will be fed later to PyGMO, therefore it must be compatible. To be PyGMO-compatible, a UDP class must have two methods:
fitness(np.array): it takes a vector of size \(n\) as input and returns a list with \(p\) values as output. \(n\) and \(p\) are respectively the dimension of the problem (in our case, \(n = 2\)) and the number of objectives (in our case, \(p = 1\) because it is a single-objective optimization).get_bounds(): it takes no input and returns a tuple of two \(n\)-dimensional lists, defining respectively the lower and upper boundaries of each variable. The dimension of the problem (i.e. the value of \(n\)) is automatically inferred by the return type of the this function.
import math
class HimmelblauOptimization:
"""
This class defines a PyGMO-compatible User-Defined Optimization Problem.
"""
def __init__(self,
x_min: float,
x_max: float,
y_min: float,
y_max: float):
"""
Constructor for the HimmelblauOptimization class.
"""
self.x_min = x_min
self.x_max = x_max
self.y_min = y_min
self.y_max = y_max
def get_bounds(self):
"""
Defines the boundaries of the search space.
"""
return ([self.x_min, self.y_min], [self.x_max, self.y_max])
def fitness(self,
x: list):
"""
Computes the fitness value for the problem.
"""
function_value = math.pow(x[0] * x[0] + x[1] - 11.0, 2.0) + math.pow(x[0] + x[1] * x[1] - 7.0, 2.0)
return [function_value]
See also
For more information, see the PyGMO documentation about defining an UDP class.
2. Creation of a PyGMO problem
Once the UDP class is created, we must create a PyGMO problem object by passing
an instance of our class to pygmo.problem. Note that an instance of the UDP class
must be passed as input to pygmo.problem() and NOT the class itself. It is also possible to use a PyGMO UDP, i.e.
a problem that is already defined in PyGMO, but it will not be shown in this tutorial. In this example,
we will use only one generation. More information about the PyGMO problem class is available
on the PyGMO website.
import pygmo
# Instantiation of the UDP problem
udp = HimmelblauOptimization(-5.0, 5.0, -5.0, 5.0)
# Creation of the pygmo problem object
prob = pygmo.problem(udp)
3. Selection of the algorithm
Now we must choose a specific optimization algorithm to be passed to pygmo.algorithm. For this example, we will use
the Differential Evolution algorithm (DE). Many different algorithms are available
through PyGMO, including heuristic methods and local optimizers. It is also possible to create a User-Defined Algorithm
(UDA), but in this tutorial we will use an algorithm readily available in PyGMO. Since the algorithm internally uses a
random number generator, a seed can be passed as an optional input argument to ensure reproducibility.
See also
For more information, see the PyGMO documentation about available algorithms and the PyGMO algorithm class.
Note
During the actual optimization process, fixing the seed is probably what you do not want to do.
# Define number of generations
number_of_generations = 1
# Create Differential Evolution object by passing the number of generations as input
de_algo = pygmo.de(gen=number_of_generations)
# Create pygmo algorithm object
algo = pygmo.algorithm(de_algo)
4. Initialization of a population
As a final preliminary step, a population of individuals must be initialized with pygmo.population. Each individual has an associated
decision vector which can change (evolution), the resulting fitness vector, and an unique ID to allow their tracking.
The population is initialized starting from a specific problem to ensure that all individuals are
compatible with the UDP. The default population size is 0; in this example, we use 1000 individuals.
Similarly to what was done for the algorithm, since the population initialization is random,
a seed can be passed as an optional input argument to ensure reproducibility.
# Set population size
pop_size = 1000
# Set seed
current_seed = 171015
# Create population
pop = pygmo.population(prob, size=pop_size, seed=current_seed)
See also
For more information, see the page from the PyGMO documentation about the PyGMO population class.
5. Evolve the population
To actually solve the problem, it is necessary to evolve the population.
This can be done by calling the evolve() method of the pygmo.algorithm object. We do so 100 times
in a recursive manner. At each evolution stage, it is possible to retrieve the full population through the get_x()
method and, analogously, the related fitness values with get_f(). If we are only interested in the best individual
of each evolution stage, we can find its index through the pop.best_idx() method. On the contrary, the champion_x
(and the related champion_f) attributes retrieves the decision variable vector and its fitness value. Note that the
champion is the best individual across all evolutionary stages (not necessarily the best individual found at the last
evolution).
# Set number of evolutions
number_of_evolutions = 100
# Initialize empty containers
individuals_list = []
fitness_list = []
# Evolve population multiple times
for i in range(number_of_evolutions):
pop = algo.evolve(pop)
individuals_list.append(pop.get_x()[pop.best_idx()])
fitness_list.append(pop.get_f()[pop.best_idx()])
See also
For more information, see the PyGMO documentation about evolving a PyGMO population.
6. Visualization of the results
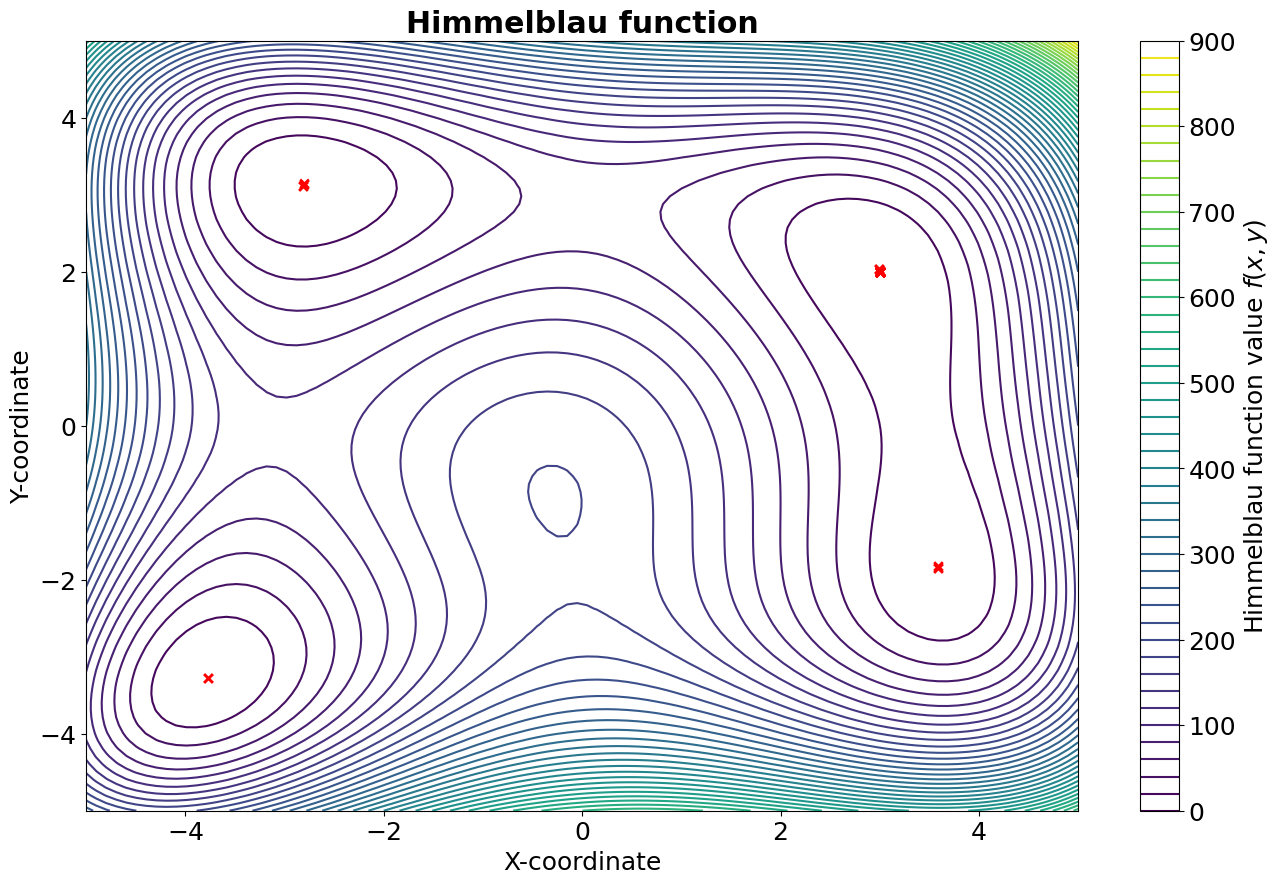
In the following figure, a contour plot of the Himmelblau’s function is reported, where the red X represent the best individuals of each generation. As it can be seen, the algorithm manages to locate all four identical minima.
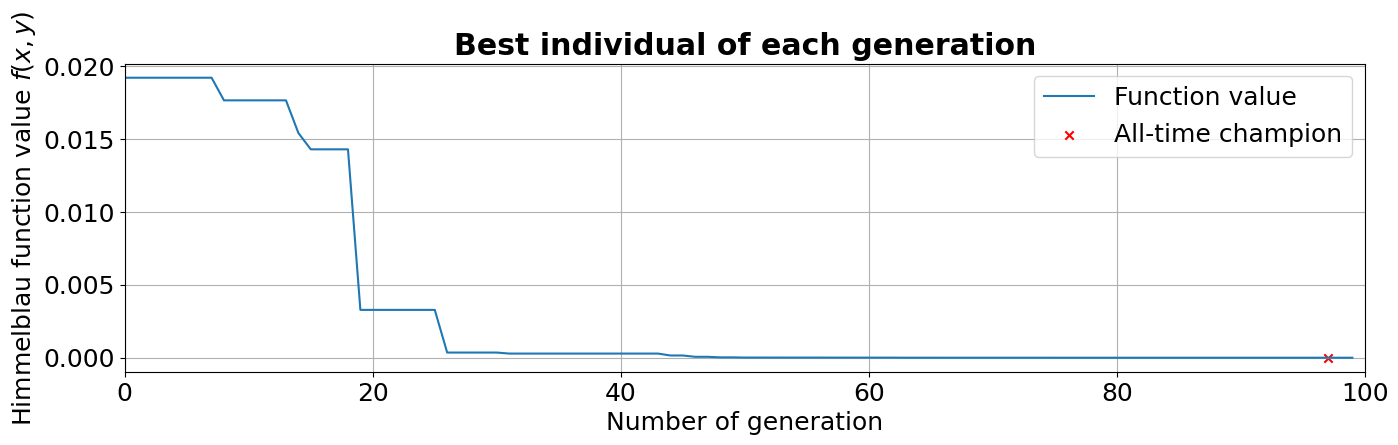
In the plot below, instead, we can see how the fitness of the best individual improves while the population is evolved. It can be seen, as anticipated before, that the champion is found a few generations before the last one.
The figure below illustrates indicatively the performance of the algorithm in the vicinity of one of the four minima.

7. Performance of the algorithm
Since this is supposed to be an introductory example, a performance analysis of the algorithm is not presented here. However, it is interesting to provide a quick comparison between the optimization conducted with PyGMO’s DE algorithm two other simple analytical methods, namely a grid search and a Monte-Carlo search (both of them were run with 1000 points per variable). In the table below, referred to the minimum located at (3,2), the results are summarized in terms of accuracy and computational expenses. As it can be noticed, the DE algorithm reaches a fitness level several orders of magnitude below the other two methods, despite using only 10% of the computational resources.
Optimization method |
Fitness value |
Decision variable difference wrt (3,2) |
Function evaluations |
PyGMO’s DE (100 gens, 1000 individuals) |
\(1.292 \cdot 10^{-11}\) |
\((-6.365 \cdot 10^{-7}, +2.382 \cdot 10^{-7})\) |
\(1.01 \cdot 10^{5}\) |
Grid search (1000 points per variable) |
\(4.215 \cdot 10^{-4}\) |
\((-2.002 \cdot 10^{-3}, -3.003 \cdot 10^{-3})\) |
\(1.00 \cdot 10^{6}\) |
Monte-Carlo search (1000 points per variable) |
\(7.095 \cdot 10^{-4}\) |
\((+4.595 \cdot 10^{-3}, -9.645 \cdot 10^{-4})\) |
\(1.00 \cdot 10^{6}\) |
- Biscani2020
Biscani et al., (2020). A parallel global multiobjective framework for optimization: pagmo. Journal of Open Source Software, 5(53), 2338, https://doi.org/10.21105/joss.02338.
Parallelization with PyGMO
In this section, a short guide is given on the parallelization of tasks in Python, and specifically for application with PyGMO. Parallelization is very useful for optimization problems, because optimizations are generally quite resource intensive processes, and this can be curbed by applying some form of parallelity. There are two flavors of parallelity in PyGMO: One utilizing multi-threading, presented in Multi-threading with Batch Fitness Evaluation and one utilizing multi-processing, presented in Multi-processing with Islands. For a more general guide on parallelization, and specifically so-called batch Fitness Evaluation (BFE), have a look at Parallelization with Python.
Multi-threading with Batch Fitness Evaluation
Multi-threading in PyGMO is used with BFE; simply evaluating some fitness function in a batch, similar to the example
explained above. For this, PyGMO has classes and methods to help setup a multi-threaded optimization. For this section,
code snippets are shown below from the hodographic shaping MGA trajectory example and adapted
to showcase the parallel capabilities. You can either define your own User-Defined Batch Fitness Evaluator (UDBFE),
explained on the pygmo documentation, or use the batch_fitness() method.
Here, the latter is explained and used as this follows more naturally from 1. Creation of the UDP class above. A
UDBFE can be applied to any UDP – with some constraints, making it more general and easier to apply out-of-the-box.
However, using UDBFE’s does not give you any control to determine how the batch is evaluated. For this reason,
batch_fitness() is used for this example.
In a UDP, BFE can be enabled by adding a batch_fitness() method to the class, as seen below. This method receives as
input a 1D flattened array of all the design parameter vectors – the first vector ranges from index [0, n], the second
from [n, 2n] and so on, with a design variable vector of length n. The output is constructed analogously, where the
length n is equal to the number of objectives. The batch_fitness() method is somewhat of a wrapper for the
fitness() method, all it has to do is convert the input array into a list of lists, then create a pool of worker
processes that can be used.
Required Show/Hide
from tudatpy.kernel.trajectory_design import shape_based_thrust, transfer_trajectory import numpy as np from typing import List, Tuple import pygmo as pg import matplotlib.pyplot as plt import multiprocessing as mp # Tudatpy imports import tudatpy from tudatpy.util import result2array from tudatpy.kernel import constants from tudatpy.kernel.numerical_simulation import environment_setup
def batch_fitness(self,
design_parameter_vectors: np.ndarray) -> List[float]:
"""
Function to evaluate the fitness. A single-objective optimization is used, in which the objective is the deltaV
necessary to execute the transfer.
"""
# Compute the final index of each type of parameters
time_of_flight_index = 3 + self.no_of_legs
incoming_velocity_index = time_of_flight_index + self.no_of_swingbys
swingby_periapsis_index = incoming_velocity_index + self.no_of_swingbys
shaping_free_coefficient_index = swingby_periapsis_index + self.total_no_shaping_free_coefficients
revolution_index = shaping_free_coefficient_index + self.no_of_legs
len_single_dpv = revolution_index
dpvs = design_parameter_vectors.reshape(len(design_parameter_vectors)//len_single_dpv, len_single_dpv)
inputs, fitnesses = [], []
for dpv in dpvs:
inputs.append([list(dpv)])
# cpu_count = len(os.sched_getaffinity(0))
cpu_count = mp.cpu_count()
with mp.get_context("spawn").Pool(processes=int(cpu_count-4)) as pool:
outputs = pool.map(self.fitness, inputs)
for output in outputs:
fitnesses.append(output)
return fitnesses
// required include statements
#include <tudat/simulation/simulation.h>
// required using-declarations
using tudat::simulation_setup;
using tudat;
Now that we have the batch_fitness() method defined, it must be called during the optimisation, which leads us to the
next code snippet. Here, we use the pygmo.set_bfe() method of a pygmo.algorithm() object to add the batch fitness
evaluation to the optimisation. Then, by default, the UDBFE pygmo.default_bfe() is given, but if the batch_fitness()
method exists in the UDP, this will automatically be used instead of the pygmo.default_bfe(). You can also use the
b keyword argument for pygmo.island and pygmo.population to add a UDBFE or an instance of pygmo.bfe, but
this is not considered here.
Required Show/Hide
from tudatpy.kernel.trajectory_design import shape_based_thrust, transfer_trajectory import numpy as np from typing import List, Tuple import pygmo as pg import matplotlib.pyplot as plt import multiprocessing as mp # Tudatpy imports import tudatpy from tudatpy.util import result2array from tudatpy.kernel import constants from tudatpy.kernel.numerical_simulation import environment_setup
if __name__ == "__main__":
bfe = True
seed = 42
pop_size = 500
# Create Pygmo problem
transfer_optimization_problem = MGAHodographicShapingTrajectoryOptimizationProblem(
central_body, transfer_body_order, bounds, departure_semi_major_axis, departure_eccentricity,
arrival_semi_major_axis, arrival_eccentricity)
prob= pg.problem(transfer_optimization_problem)
# Create algorithm and define its seed
algo = pg.gaco()
if bfe:
algo.set_bfe(pg.bfe())
algo = pg.algorithm(algo)
bfe_pop = pg.default_bfe() if bfe else None
pop = pg.population(prob=prob, size=pop_size, seed=seed)
num_gen = 150
# Initialize lists with the best individual per generation
list_of_champion_f = [pop.champion_f]
list_of_champion_x = [pop.champion_x]
# mp.freeze_support() needs to be called when using multiprocessing on windows
# mp.freeze_support()
for i in range(num_gen):
print(f'Evolution: {i+1} / {num_gen}', end='\r')
pop =algo.evolve(pop)
# Save current champion
list_of_champion_x.append(pop.champion_x)
list_of_champion_f.append(pop.champion_f)
print('Evolution finished')
// required include statements
#include <tudat/simulation/simulation.h>
// required using-declarations
using tudat::simulation_setup;
using tudat;
To show that the batch fitness evaluation actually works well, a few tests are done with various complexities: a EJ and EMEJ transfer with two different generation counts and population sizes each. Normally, the number of function evaluations would be a good indication of runtime complexity, however using BFE does not change that number. CPU time and depending on the CPU usage indirectly also clock time can give an indication of the effectiveness. It should be noted that this is software and hardware dependent, so the results should be taken with a grain of salt. For the simple problem (EJ) with few generations, adding the BFE actually increases the CPU time by almost 200%. As for the test with more generations, the addition of BFE increased run-time with almost 90%. The complex problem (EMEJ) shows slightly different behaviour; the test with few generations does improves when adding BFE by about 50%. This gain increases significantly for the test with more generations; an 80% decrease in clock time.
Note
These simulations are tested on macOS Ventura 13.1 with a 3.1 GHz Quad-Core Intel Core i7 processor only. Four cores (CPU’s) are used during the BFE.
Transfer Sequence |
Gen count and Pop size |
Batch Fitness Evaluation |
CPU time [s] |
CPU usage [-] |
Clock time [s] |
|---|---|---|---|---|---|
EJ |
gen30pop100 |
no |
17.6 |
106% |
16.7 |
yes |
130.7 |
443% |
29.5 |
||
gen300pop1000 |
no |
4500 |
78% |
5770 |
|
yes |
3000 |
405% |
735 |
||
EMEJ |
gen30pop100 |
no |
70.1 |
97% |
72.3 |
yes |
159.2 |
428% |
37.2 |
||
gen300pop1000 |
no |
4440 |
60% |
7440 |
|
yes |
5946 |
404% |
1470 |
Multi-processing with Islands
This section presents multi-processing with PyGMO using the pygmo.island and/or pygmo.archipelago class. An
island is an object that enables asynchronous optimization of its population. An archipelago is a network that connects
multiple islands – pygmo.island objects – with a pygmo.topology object. Islands can exchange individuals with
one another through this topology. This topology can configure the exchange of individuals between islands in an
archipelago. By default, the topology is the pygmo.unconnected type, which has no effect, resulting in a simple
parallel evolution.
In the code snippet below, inspired by the hodographic shaping MGA trajectory example but
parallelized with an archipelago, a group of islands evolve in parallel. Specifically, a pygmo.archipelago object is
created that initializes a number_of_islands number of pygmo.island objects using the provided algo,
prob, and pop_size arguments. pygmo.archipelago then has an evolve() method that in turn calls the
evolve() method of all pygmo.island objects separately, and allocates a python process from a process pool to
each island. The wait_check() method makes every island wait until all islands are done executing, which is needed
for any topology to exchange individuals.
Required Show/Hide
from tudatpy.kernel.trajectory_design import shape_based_thrust, transfer_trajectory import numpy as np from typing import List, Tuple import pygmo as pg import matplotlib.pyplot as plt import multiprocessing as mp # Tudatpy imports import tudatpy from tudatpy.util import result2array from tudatpy.kernel import constants from tudatpy.kernel.numerical_simulation import environment_setup
if __name__ == "__main__":
seed = 42
pop_size = 1000
number_of_islands = 4
# Create Pygmo problem
transfer_optimization_problem = MGAHodographicShapingTrajectoryOptimizationProblem(
central_body, transfer_body_order, bounds, departure_semi_major_axis, departure_eccentricity,
arrival_semi_major_axis, arrival_eccentricity)
problem = pg.problem(transfer_optimization_problem)
# Create algorithm and define its seed
algorithm = pg.algorithm(pg.sga(gen=1))
algorithm.set_seed(seed)
# Create island
archi = pg.archipelago(n=number_of_islands, algo=algorithm, prob=transfer_optimization_problem, pop_size=pop_size)
num_gen = 40
# Initialize lists with the best individual per generation
list_of_champion_f = []
list_of_champion_x = []
# mp.freeze_support() needs to be called when using multiprocessing on windows
# mp.freeze_support()
for i in range(num_gen):
print('Evolution: %i / %i' % (i+1, num_gen))
archi.evolve() # Evolve archi
archi.wait_check() # Wait until all evolution tasks in the archi finish
# Save current champion
list_of_champion_x.append(archi.get_champion_x())
list_of_champion_f.append(archi.get_champion_f())
print('Evolution finished')
// required include statements
#include <tudat/simulation/simulation.h>
// required using-declarations
using tudat::simulation_setup;
using tudat;